The goals of our research are collectively focused on defining the genetic pathways and transcriptional signatures that govern cell lineage commitment. The knowledge obtained in our research is applicable toward developing directed stem cell differentiation strategies for advancing cell-based therapeutics and increasing our understanding of disease mechanisms. We utilize platforms in developmental embryology, single-cell transcriptomic and proteomic analyses, embryonic and induced pluripotent stem cell culture and characterization, and the lineage-specific differentiation of pluripotent stem cells to define and investigate novel genetic correlations. We maintain active industry collaborations using high throughput single-cell microfluidic platforms to examine the transcriptional profiles of individual cells in developing systems, and we utilize genome-wide analyses for investigating cell lineage commitment at single-cell resolution. The primary goal of our studies is the mapping of cell lineages at high resolution based on gene expression profiles and the ultimate contributions of our research will define the genetic regulatory mechanisms which guide developmental processes and specify cell types. Our incorporation of high throughput technologies for whole transcriptome analyses and correlations with epigenetic and proteomic states provide a foundation for detailing stages of development at a resolution previously unattainable. Our current research platform sets a stage for the novel discovery of well-defined cellular phenotypes and provides a resource for the future investigation of disease models to increase our understanding of the precise control of cell lineage specification during development.
Gene correlation network generated from whole-embryo, whole-transcriptome single-cell RNA sequencing
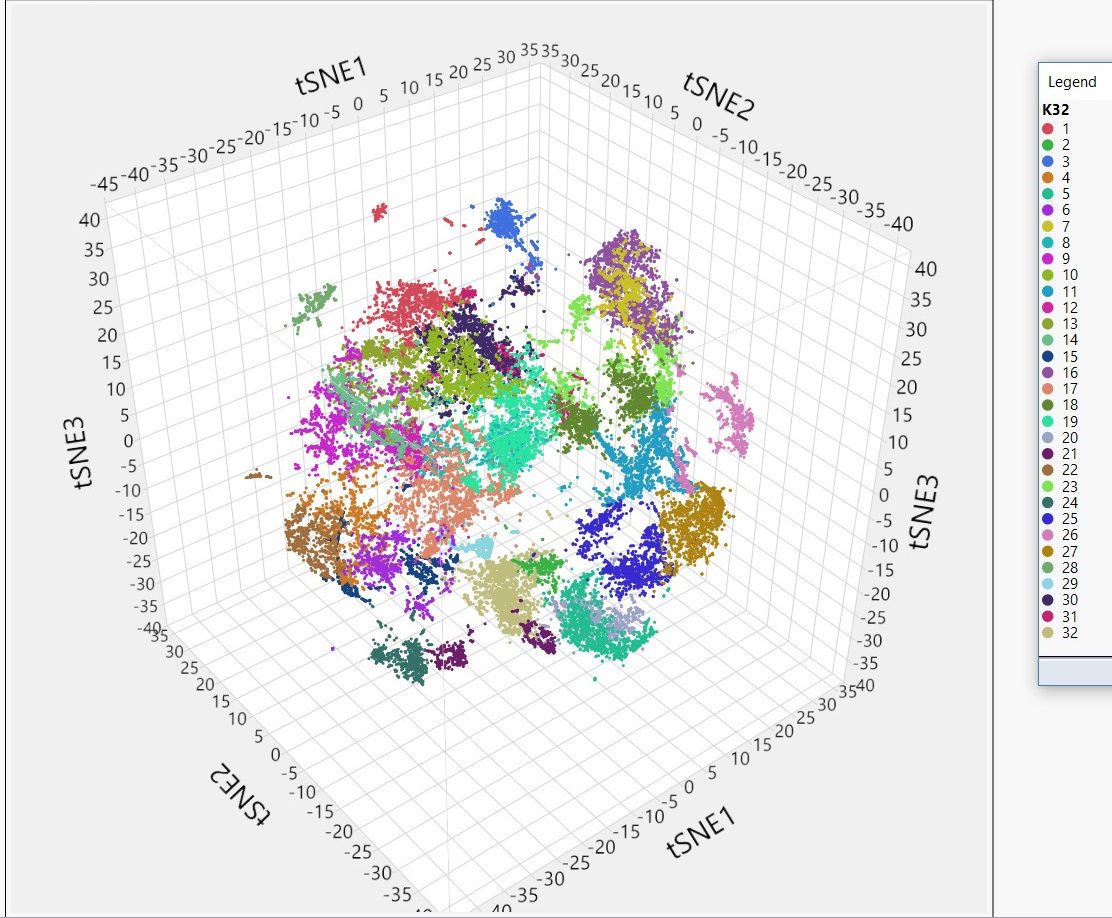
Clustering of cells from the developing mouse embryo based on transcriptional similarity from single-cell RNA sequencing